Abba Musa Abdullahi*
Neurology/Neuroscience, Hasiya Bayero Paediatric Hospital, Kano, NGA
*Corresponding author: Abdullahi AM, MBBS, MPH, Neurology/Neuroscience, Hasiya Bayero Paediatric Hospital, Kano, NGA
Abstract
Coronavirus disease of the 2019 (COVID-19) is a contagious disease with a varying range of severity ranging from mild to moderate, severe, and critical. It has devastating consequences on the healthcare system, economy, and political system making it a public health emergency. The viral aetiologic agent was named by the International Committee on Taxonomy of Virus (ICTV) as SARS-CoV-2. It was first discovered in the Wuhan city of Hubei province, China in mid-December from where it spread to every continent on earth. The viral envelope is nearly spherical with crown-like surface projections known as spikes which aid in viral attachment to the hosts’ tissues. It has a single-stranded RNA genome with many nucleotides bases encoding thousands of amino acids with two adjoining untranslated regions (UTRs) and one long open reading frame (ORF) encoding polyproteins. The main route of transmission is by a human-to-human transmission via direct contact or via respiratory droplets spread by coughing, sneezing, or talking from an infected individual with an average incubation period of 4-6 days, however, 14 days incubation period was suggested by experts especially during the quarantine. As of June 2, 2020, the global confirmed cases were reported by the European Centre for Disease Control and Prevention (ECDC) as 6,245,352 cases with global death was recorded as 376,427 deaths.
Keywords: coronavirus, COVID-19, SARS-CoV-2, virology, genome, virion, spikes, epidemiology
1. Introduction
The term coronavirus (CoV) was derived from the Latin name ‘corona’ which means the crown-like spikes or halo around the viral envelop viewed under the electron a microscope and belongs to the family coronaviridae [1]. Coronaviruses have wide range of natural hosts and are believed to be the largest group of positive-sense RNA viruses causing an increasing threat to public health worldwide with devastating economic impacts [2]. The virus was first isolated from chickens by Beaudette and Hudson in 1937 which was termed as infectious bronchitis virus (IBV) [3], and in 1946 a transmissible gastroenteritis in swine was identified which was then followed by recovery of murine hepatitis viruses (MHV) in 1949 by Cheever and colleagues [4]. The human coronaviruses (HCoVs) were first discovered in 1965 by Tyrrell and Bynoe from nasal washings of a male child, which led to the proper categorization of the coronavirus genus [5]. These viruses are commonly found in bats, however, intermediate hosts like livestock and household pets are identified to be responsible for emergence and transmission of these viruses from bats to humans [6], and knowledge of bat origin of human coronaviruses could have a tremendous impact in predicting and prevention of possible occurrence of the coronavirus-related pandemic [7].
2. Virology
2.1 Overview
Generally, most coronaviruses are similar in structures which are enveloped but nearly spherical with a diameter of about 80-220nm. They are marked by prominent crown-like surface projections (spikes), 20nm in length, made up of heavily glycosylated type I glycoprotein, called spike protein (S) [10]. They are positive-sense single-stranded RNA viruses with a nucleocapsid of helical symmetry [6]. The virion envelope is made of inner and outer shells separated by a translucent space with a tongue-like inner membrane. Additionally, the virion has an internal ribonucleoprotein (RNP) measuring 1-2nm in length which is condensed into coiled structures [10]. The genome is composed of a single-stranded RNA linear molecule which is capped, polyadenylated and infectious harboring about 15000 to 20000 nucleotides with many genes encoding the polymerase and structural proteins a well as several open reading frames (ORFs) interspersed between these genes which encode non-structural proteins [11].
This genome is organized into 6-7 regions separated by junction sequences which contain signals for transcriptions of several mRNAs and each region contains one or more ORFs. There are three major structural proteins in the coronavirus virion: a nucleocapsid protein (N), a small integral membrane glycoprotein (M) and a large spike glycoprotein (S), however, additional proteins are found in some coronaviruses [12]. The N-protein play an important role in the encapsidating of genomic RNA, its subsequent incorporation into the virion through the formation of RNP and replication of RNA; M-protein facilitates virus maturation and determine the site for viral particles assembly; S-protein play a role in receptor recognition and membrane fusion which facilitate entry into the host cells [13].
The whole genome sequence of Covid-19 showed that it is a member of β-CoVs of group 2B with 79.0 % and 51.8 % similarity with SARS- CoV and MERS - CoV, respectively [14]. The SARS-CoV is a positive-sense single-stranded ribonucleic acid (RNA) virus consisting of thousands of nucleosides bases which phylogenetically related to other human coronaviruses especiallyHCoV-OC43 and HCoV-229E [15]. The genome of the SARS-CoV is made up of basically five main open reading frames (ORFs) which encode the replicase polyprotein; the spike (S), envelope (E), and membrane (M) glycoproteins; and the nucleocapsid protein (N) that are structurally and functionally similar to proteins of other coronaviruses. It also has a transcription regulating sequence (TRS) located near the beginning of each ORF and at the 3’end which discontinue RNA transcription and therefore facilitate syntheses of subgenomic negative-sense RNA species which may be translated in infected cells [16] (Figure 1).
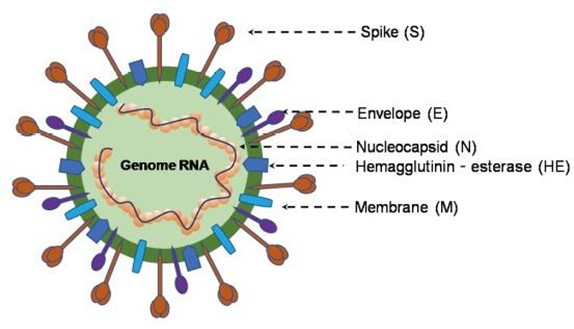
Figure 1: structure of β-Coronavirus genome (Adapted with modification from Jin et al. (2020):Virology, Epidemiology, Pathogenesis, and Control of COVID-19)
2.2 Source and Genome Characterization
Genome sequencing of samples from bronchoalveolar lavage fluid and cultured isolates in late December 2019 from nine patients presented to Wuhan hospitals with symptoms of viral pneumonia who were tested negative for common causes of pneumonia revealed similarity among the samples with over 99.9 % sequence identity, closely resembling SARS-CoV (about 79 %) and MERS-CoV (about 50 %) but not identical [17], resulting in the viral identification by WHO as 2019-nCoV [9]. However, due to its close but a relatively distant relationship with SARS-CoV and MERS-CoV, the Coronavirus Study Group (CSG) categorized the virus pathogen as SARS-CoV-2 and the disease entity it caused was named by WHO as COVID-19 [18].
The origin of SARS-CoV-2 is debatable. Many attributed the source to bats due to 96 % similarity of the SARS-CoV-2 genome at the whole-genome level with bat-derived coronavirus genome [19]. In a study by Zhang and colleagues, Pangolin-CoV at the whole-genome level is found to be 91.02 % identical to SARS-CoV-2 indicating the probable Pangolin origin of SARS-CoV-2 and this is supported by the consistency of the five main amino acids in the RBD between Pangolin-CoV and SARS-CoV-2 [20]. However, a unique RRAR motif in the spike protein of human SARS-CoV-2 that is not present in coronaviruses isolated from pangolins indicates that SARS-CoV-2 may not come directly from pangolins [21]. Additionally, some strains of SARS-CoV-2 were isolated a few months earlier before COVID-19 was officially reported, thus disputing bats as a potential origin [22]. Some pieces of evidence from phyloepidemiological approaches are suggesting the possibility of importation of SARS- CoV-2 from other places to the Huanan Seafood Market [23]. Similarly, the virus was shown to be a recombinant virus between the bat coronavirus and another coronavirus with unknown origin with a snake as the most likely animal reservoir [9].
SARS-CoV-2 was found to be single-stranded RNA genome with 29891 nucleotides bases encoding 9860 amino acids with two adjoining untranslated regions (UTRs) and one long ORF encoding polyproteins [20]. The genome has six main ORFs with many more accessory genes with a 29,891-base-pair CoV genome [19]. As with other human coronaviruses, the SARS-CoV-2 genome has a transcription regulatory sequences and two terminal untranslated regions which facilitate the synthesis of subgenomic mRNA expressed ORFs [24]. Additionally, the genome has 5’ and 3’ terminal sequences (265 nt at the 5’ terminal and 229 nt at the 3’ terminal region) resembling other human coronaviruses with 5’ replicase open reading frame (ORF) 1ab-S-envelope(E)- membrane(M)-N-3’ gene order (Jin et al., 2020). The genome has a diameter of about 60-100nm with a length of about 29.9 kb [25]. It is unstable and can easy be deactivated by UV or when heated at 56C for at least 30 min, and sensitive to most disinfectants such as diethyl ether, 75 % ethanol, chlorine, peracetic acid, and chloroform [26]. Three deletions were found in isolates from Japan, USA and Australia with ninety-three mutations found over the entire genomes. Additionally, eight mutations were found in the spike surface glycoprotein, particularly three mutations (D354, Y364, and F367) found in the spike surface glycoprotein a receptor-binding domain which may indicate an important revolutionary trend of SARS-CoV- 2 in the future [21] (Figure 2).
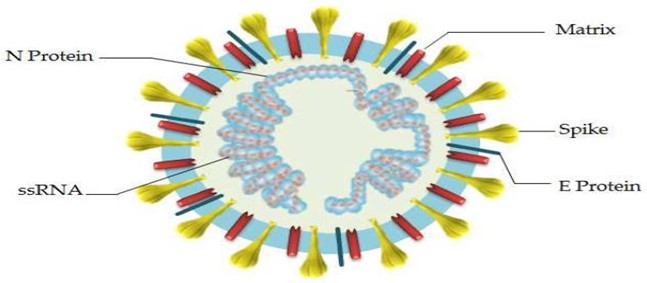
Figure 2: SARS-CoV-2 Structure (adapted from Astuti and Ysrafil (2020): Severe Acute Respiratory Syndrome Coronavirus 2 (SARSCoV-2): An Overview of Viral Structure and Host Response).
2.3 Virion Proteins and Receptor Interaction
The virion has four main structural proteins: The Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) proteins as in other human coronaviruses but differentially deficient in the hemagglutinin- esterase gene [27]. The spike (S) protein is made up of two subunits, S1 and S2. The former consists of a signal peptide with two domains: an N-terminal domain (NTD) and a receptor-binding domain (RBD); whereas the latter consists of conserved fusion peptide (FP), heptad repeat (HR) 1 and 2, transmembrane domain (TM), and a cytoplasmic domain (CP) [28]. The similarity between S1 and S2 subunits with both the two bats SARS-like CoVs (SL-CoV ZXC21 and ZC45) and human SARS-CoV is found to be 70% and 99% respectively making the S2 subunit as an important target for both preventive and therapeutic approach against SARS-CoV-2 [29,30]. The receptor- binding domain (RBD) of S1 subunit is made up of two subdomains: external and internal subdomains. The former harbors most of the amino acid differences of RBD and is the facilitator for the direct viral interaction with the host receptor, human angiotensin-converting enzyme 2 (ACE2); while the latter is highly conserved [29]. (Figure 3).
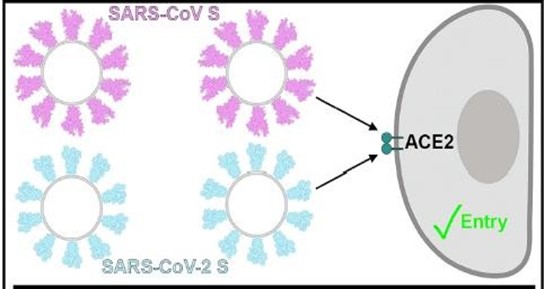
Figure 3: Receptor interaction (Adopted with modification from Walls et al., 2020: structure, function and antigenicity of the SARS-CoV-2 glycoproteins).
The Orf3b and Orf8 are accessory proteins that are found in human coronaviruses that have also been identified in SARS-CoV-2 with some peculiarities. The Orf3b the protein of SARS-CoV-2 encodes a short protein with a putative binding site for Karyopherin (K) proteins which impair the interferon signaling network and the host innate defense by binding to the K-proteins, a function resembling SARS-CoV Orf6 protein function [24]. Contrary to the Orf8 of other coronaviruses, the SARS-CoV-2 Orf8 does not contain any functional domain due to lack of VLVVL (amino acid 75–79) motif found in SARS-CoV Orf8b in which it stimulates intracellular stress pathways with activation of NLRP3 inflammasomes [29]. With additional study, SARS-CoV-2 was demonstrated to use the same cell entry receptor, ACE2, as SARS-CoV, not CD26 as MERS-CoV. Also, structural analysis by Cryo-EM have shown that n The SARS-CoV-2 protein binds ACE2 with 10-20 folds higher affinity than SARS- CoVs indicating that, SARS-CoV-2 may be more infectious to humans than SARS-CoV [21].
2.4 Genomic Comparison
The whole level sequence analysis of SARS-CoV-2 revealed that it shares 79 % sequence identity to SARS-CoV and around 50 % to MERS-CoV2. However, the seven conserved replicase domains in ORF1ab (used for CoV species classification) of SARS-CoV-2 shows 94.6 % similarity to SARS-CoV, meaning that the two belong to same species. The receptor-binding protein spike (S) gene of SARS-CoV- 2 is highly divergent to all previously described SARSr-CoVs with less than 75 % sequence identity to except a 93.1 % identity to RaTG13. Homology modeling revealed SARS-CoV-2 had a similar receptor-binding domain structure to that of SARS-CoV [21]. Early sequencing the analysis revealed more than 99.9 % similarity among the samples collected, however, further studies indicate that SARS- CoV-2 has undergone mutation and substitutions at some codons evolving into two major types: L and S types with the L type exhibiting more aggressive behavior and likely to spread more quickly than the S type [26].
3 Epidemiology
3. 1 Disease Source
The main sources of infection are the COVID19 infected patients and the more severe the disease, the more contagious it is [31]. Additionally, individuals without clinical signs and symptoms of COVID19 (asymptomatics) were reported to be potential sources of infection as they were proven to shed the infectious virus [32]. Similarly, patients recovered from COVID19 were reported to have shown positive RT-PCR test again, however, it is clear whether they are still infectious and can transmit the disease [33].
3. 2 Incubation Period
The average incubation period is between 4-6days [34]. In a Wuhan study of 425 confirmed cases, the mean incubation period was found to be 5.2days [35]. In another study by Linton and colleagues, the mean incubation period was estimated to be 5days [36]. In the Chinese studies conducted outside Wuhan among 88 confirmed cases, a mean incubation of 6.4days was reported [37]. Lauer and colleagues estimated that 2.5 % of the patients will develop symptoms within 2.2 days and 97.5 % of patients will develop symptoms within 11.5 days [38]. However, an incubation period of 1-19days was reported in a familial cluster of five patients indicating that the COVID-19 incubation period is almost like MERS and SARS but slightly longer than influenza [39]. The 19days incubation period is a rare occurrence; therefore, specialists and professionals suggest the maximum incubation period as 14days for quarantine [40].
3. 3 Disease Pattern
As most other viral respiratory diseases, COVID19 is also a self- limiting disease with the majority of the cases occurring as mild diseases that can recover within 1-2weeks [41]. The disease can occur in five different patterns: asymptomatic without any clinical sign or symptom, mild with mild upper respiratory tract symptoms, moderate with symptoms of pneumonia, severe with severe symptoms of pneumonia requiring oxygen and critical with acute respiratory distress syndrome (ARDS) or respiratory failure, shock, or multiple organ dysfunction [39]. In a report published by the Chinese Center for Disease Control and Prevention, 1.2 % cases were asymptomatic, 81 % were mild cases, 14 % were severe cases, 5 % were critical cases with a case fatality rate of 2.3 % which is remarkably high in elderly and individuals with associated co-morbid conditions [42]. Healthcare workers are one of the most susceptible hosts of COVID19 due to their close contact with infected individuals. About 20 % and 19 % of healthcare workers in Italy and the United States were infected with COVID19 with the majority of the cases reported as mild cases but with significant mortalities [43].
3. 4 Route of Transmission and Susceptible Host
The main route of transmission is by a human-to-human transmission via direct contact or via respiratory droplets spread by coughing, sneezing, or talking from an infected individual resembling the mode of spread of influenza [44]. Currently, there some evidence of the virus detected in blood and stool specimen, thus serving as potential routes of transmissions [21]. Based on the available evidence, anybody, irrespective of his age, sex, or race that has close contact with an individual infected with COVID19 is susceptible to contract the disease [34]. In a study by Wu and colleagues, the sex distribution was 1.06:1 indicating that the disease affects both females and males almost equally. However, the age distribution was remarkable with most of the cases (87 %) affecting individuals within the age range of 30-79yrs almost sparing children (1 %) and teenagers (1 %) [41]. currently, no evidence to support the vertical transmission of COVID19, however, adverse health outcomes were reported in some newborns born to COVID19 infected mothers [45]. The risks for adverse outcomes in COVID19 infected patients include increasing age and associated co-morbidities like cardiovascular disease, diabetes mellitus, chronic respiratory disease, hypertension, and cancer [43].
3. 5 Geographical Distribution
Following the early report of COVID-19 identification from mainland China, the disease rapidly spread across the continent and the number of cases increased exponentially worldwide with the first imported case reported from Thailand on early January [43]. As of May 31, 2020, the COVID-19 pandemic has affected six geographical regions: Africa, Americas, Eastern Mediterranean region, Europe, South-East Asia, and Western Pacific [46]. By June 2, 2020, the European Centre for Disease Prevention and Control (ECDC) reported the highest incidence of COVID-19 in Americas region (n = 2,956,532) with United States having the highest incidence worldwide (n = 1,811,277), followed by Europe (n = 1,975,341), Asia (n = 1,151,637) and then Africa (n = 152,485). Similarly, the number of new cases were highest is Americas region, followed by Europe, Asia and then Africa. However, the overall death rate was highest in Europe (n = 175,572), followed by Americas (n = 165,262), Asia (n = 31,110) and then Africa (4,344) with the least death recorded in Oceania (ECDC COVID-19 Situation Update Worldwide, June 2, 2020) (Figure 4).
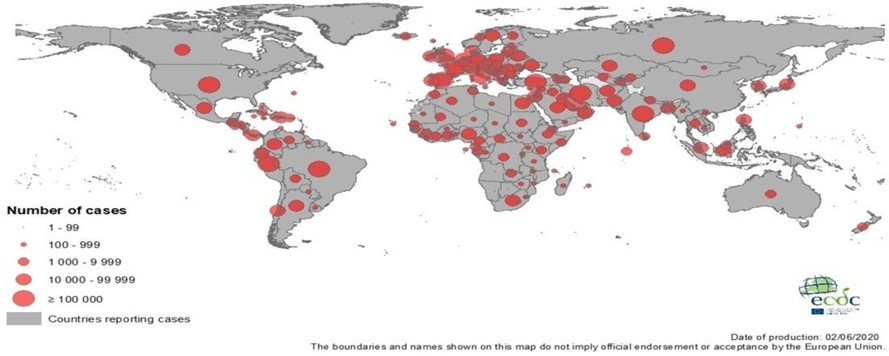
Figure 4: geographical distribution of Covid19 Worldwide, as of June 2, 2020. Adapted from ECDC Situation Update Worldwide, June 2, 2020.
3. 6 Disease Chronology
Many cases of pneumonia-like cases of unknown causes were reported in the mid-December 2019 from the central Chinese city of Wuhan [44]. Five patients presented with fever, cough, and dyspnea accompanied by complications of acute respiratory distress syndrome were hospitalized from December 18 to December 29, 2019 [47]. On December 31, 2019, China alerted WHO about a cluster of 41 patients with new cases of unusual pneumonia in Wuhan city comprising of about 11 million people located in the central Hubei province [48]. Around January 2, 2019, 41 hospitalized patients were reported to be laboratory-confirmed cases of the new virus, and most of them were males some of whom were exposed to the Huanan seafood market [49]. The World Health Organization (WHO) on January 7, 2020, identified the virus as a novel coronavirus and officially but tentatively named it as 2019-nCoV, the new coronavirus in 2019 [9]. Around January 13, 16 and 21, confirmed imported cases in three Asian countries comprising of Thailand, Japan, and Korea were respectively reported marking the beginning of the global pandemic [2]. By Jan 22, 2020, a total of 571 cases of the 2019-new coronavirus (2019-nCoV) have been reported in 25 provinces (districts and cities) in China [50]. On Jan 24, 2020, with reports of thirteen exportation events, the cumulative incidence of the new coronavirus (2019-nCoV) in China was estimated to be 5502 cases [50]. By Jan 25, 2020, a total of 1975 cases of 2019-nCoV infection were confirmed in mainland China with a total of 56 deaths that have occurred [52].
As of January 30, 2020, the confirmed cases in China were estimated to be 7734 cases and concurrently 90 more cases were also confirmed in many countries including Taiwan, Thailand, Vietnam, Malaysia, Nepal, Sri Lanka, Cambodia, Japan, Singapore, Republic of Korea, United Arab Emirates, United States, The Philippines, India, Australia, Canada, Finland, France, and Germany with a case fatality rate estimated to be 2.2 % [53]. On January 31, the WHO declared the disease as the Public Health Emergency of International Concern (PHEIC), which indicate that the disease can pose threats to many countries and therefore requires an active coordinated international response [1]. By 31 Jan 2020, the total number of confirmed cases was 9,826 cases globally affecting 19 countries with 213 mortalities [54].
On February 11, both ICTV and WHO comes up with new but different names for the virus. The former named as SARS-CoV2 while the later named it as COVID19 (WHO COVID-19 Situation Report-22, 2020). As of February 29, 2020, the total number of confirmed cases was 85,403 cases globally involving 53 countries with 2,924 global deaths [55]. On March 11, the WHO declared the disease as a “pandemic public health menace” [1] (Figure 5). By March 31, 2020, the global confirmed cases reached 750,890 cases with 36,405 global deaths [56]. A global confirmed case was reported to be 3,090,445 cases on April 30, 2020 with 217,769 total deaths [57]. As of May 31, 2020, the total confirmed cases globally were 5,934,936 cases with 367,166 global deaths [46]. As of June 2, 2020 (the last day of preparing this manuscript), the global confirmed cases were reported by ECDC as 6,245,352 cases with global deaths recorded as 376,427 deaths (ECDC COVID-19 Situation Update Worldwide, June 2, 2020) (Table 1).
Table 1: Shows the Global Incidence of COVID-19 across Countries with the Case Fatality Rate from December to May, 2020
|
Date |
Number of Confirmed Cases |
Deaths |
Case Fatality Rate (%) |
|
December 31, 2019 |
41 |
0 |
0 |
|
January 31, 2020 |
9826 |
213 |
2.2 |
|
February 29, 2020 |
85,403 |
2924 |
3.4 |
|
March 31, 2020 |
750,890 |
36,405 |
4.8 |
|
April 30, 2020 |
3,090,445 |
217,769 |
7.0 |
|
May 31, 2020 |
5,934,936 |
367,166 |
6.2 |
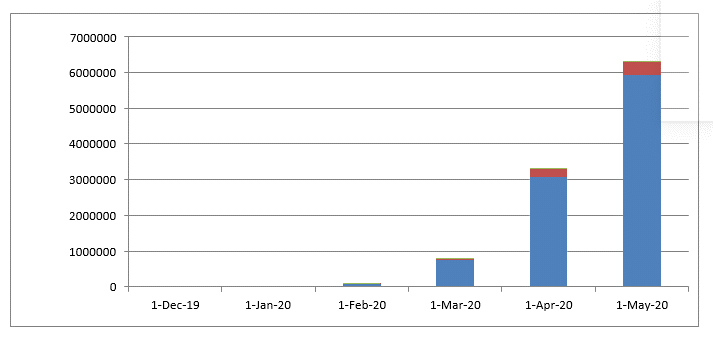
Figure 5: chart demonstrating the number of confirmed cases and deaths from Dec 31, 2019 to May 31, 2020.
4. Conclusion
SARS-CoV-2 is an emerging disease of public health importance. The majority of the cases are mild self-limiting conditions but few cases present as serious and critical conditions requiring high input of clinical management. It is highly contagious with devastating consequences especially in the elderly and those with associated medical conditions like diabetes, hypertension, respiratory diseases, or cardiovascular conditions. The primary route of transmission is through contact directly with an infected person or via respiratory droplets with an average incubation period of 4-6 days. The structure of the virus resembles SARS-CoV and MERS-CoV in many aspects but appeared to have some peculiarities with bats as the most probable source of the disease. It has a rapid geographical distribution making it a global pandemic and serious infectious disease requiring urgent and active measures to curtail its spread. It is clear that the Wuhan Seafood market was not the only origin of this disease indicating other potential origins as was identified in this paper. Effective control measures, appropriate preventive strategies, and active case surveillance are the best approaches to curtail the spread of this virus and to win the battle against it.
Conflict of Interest: None to disclose
Funding: None
References
- Jebril N (2020) World Health Organization declared a pandemic public health menace : A World Health Organization declared a pandemic public health menace : A systematic review of the coronavirus disease 2019 “ COVID - 19 ”, up to 26 th March 2020.
- Li G, Fan Y, Lai Y, Han T, Wang W, et al. (2020) Coronavirus infections and immune responses. J Med Virol 424-432.
- Wevers BA, van der Hoek L (2009) Recently Discovered Human Coronaviruses. Clin Lab Med 29(4): 715-724.
- Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S (2019) Coronaviruses. (M): 488-523.
- Myint SH (1995) Human Coronavirus Infections. The Coronaviridae. (Table I) : 389-401.
- Khan S, Siddique R, Shereen A, Ali A, Liu J, et al. (2020) crossm Emergence of a Novel Coronavirus , Severe Acute Respiratory Syndrome Coronavirus 2 : Biology and Therapeutic Options. 1-9.
- Hu B, Ge X, Wang L, Shi Z (2015) Bat origin of human coronaviruses. Virol J 12: 1-10.
- Gorbalenya AE, Baker SC, Baric RS, Groot RJ De, Gulyaeva AA, Haagmans BL, et al. The species and its viruses – a statement of the Coronavirus Study Group. 2020.
- Cheng ZJ, Shan J (2020) 2019 Novel coronavirus : where we are and what we know. Infection 48: 155-163.
- Lai MMC, Cavanaght D (1997) The Molecular Biology Of Coronaviruses Adv Virus Res 48: 1-100.
- W Spaan, D Cavanagh, M C Horzinek (1989) Coronaviruses : Structure and Genome Expression 69(12): 2939-2952.
- Siddell SG (1995) The Coronaviridae. The Coronaviridae 1-10.
- Li F (2016) Structure , Function , and Evolution of Coronavirus Spike Proteins 3: 237-264.
- Park SE (2020) Epidemiology , virology , and clinical features of severe acute respiratory syndrome -coronavirus-2 ( SARS-CoV- 2 ; Coronavirus Disease-19 ). 63(4): 119-124.
- Satija N, Lal SK (2007) The molecular biology of SARS coronavirus. Ann N Y Acad Sci 1102(1): 26-38.
- Holmes K V, Enjuanes L (2003) The SARS Coronavirus: A Postgenomic Era. Science 300: 1377-1378.
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224): 565-574.
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, et al. (2020) The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV- 2. Nat Microbiol. 5(4): 536-544.
- Zhou P, Yang X Lou, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798): 270-273.
- Zhang T, Wu Q, Zhang Z (2020) Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol 30(7): 1346-1351.e2.
- Wang Y, Wang Y, Chen Y, Qin Q (2020) Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol 92(6): 568-576.
- Xiong C, Jiang L, Chen Y, Jiang Q (2020) Evolution and variation of 2019-novel coronavirus.
- Yu W, Tang G, Zhang L, Corlett RT (2020) Decoding the evolution and transmissions of the novel using whole genomic data 41(3): 247-257.
- Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9(1): 221-236.
- Astuti I, Ysrafil (2020) Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diab Metab Synd: Clin Res Rev 14(4): 407-412.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C, et al. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483): 1263: 1260-1263.
- Wu F, Zhao S, Yu B, Chen Y, Wang W, et al. (2020) Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China.
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2 Science 367(6485): 1444-1448.
- Walls AC, Park Y, Tortorici MA, Wall A, Mcguire AT, et al. (2020) Structure , Function ,and Antigenicity of the SARS-CoV- 2 Spike Glycoprotein. Cell 1-12.
- Gorbalenya AE, Krupovic M, Mushegian A, Kropinski AM, Siddell SG, et al. (2020) The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nature Microbiology 5(5): 668-674.
- Chan JFW, Yuan S, Kok KH, To KKW, Chu H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395(10223): P514-523.
- Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, et al. (2020) Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med 382(13): 1278-1280.
- Lan L, Xu D, Ye G, Xia C, Wang S, et al. (2020) Positive RT- PCR Test Results in Patients Recovered from COVID-19. JAMA. J Am Med Assoc 323(15): 1502-1503.
- Park M, Cook AR, Lim JT, Sun Y, Dickens BL (2020) A Systematic Review of COVID-19 Epidemiology Based on Current Evidence. J Clin Med 9(4): 967.
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus- infected pneumonia. N Engl J Med 382(13): 1199-1207.
- Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, et al. (2020) Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data. J Clin Med 9(2): 538.
- Backer JA, Klinkenberg D, Wallinga J (2020) Incubation period of 2019 novel coronavirus (2019- nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Eurosurveillance 25(5): 1-6.
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, et al. (2020) The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: Estimation and application. Ann Intern Med 172(9): 577-582.
- Bulut C, Kato Y (2020) Epidemiology of covid-19. Turkish J Med Sci 50(SI-1): 563-570.
- Zhai P, Ding Y, Wu X, Long J, Zhong Y, et al. (2020) The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents. 55(5): 105955.
- Wu Z, McGoogan JM (2020) Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA - J Am Med Assoc 323(13): 1239-1242.
- Surveillances V (2020) The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronavirus Diseases ( COVID-19 ) — China. 2(x): 1-10.
- Chowdhury SD, Oommen AM (2020) Epidemiology of COVID- 19. J Dig Endosc 11(1): 3-7.
- Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109: 102433.
- Zhu H, Wang L, Fang C, Peng S, Zhang L, et al. (2020) Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl Pediatr 9(1): 51-60.
- WHO (2020) Coronavirus disease 2019 ( COVID-19 ) Situation Report - 132.
- Ren L, Wang Y, Wu Z, Xiang Z, Guo L, et al. (2020) Identification of a novel coronavirus causing severe pneumonia in human : a descriptive study. Chin Med J (Engl) 133(9): 1015- 1024.
- Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, et al. (2020) World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg 76: 71-76.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan , China. Lancet 395(10223):497-506.
- WHO (2020) Novel Coronavirus ( 2019-nCoV ). Situation Report-22.
- Nishiura H, Jung S, Linton NM, Kinoshita R, Yang Y, et al. The Extent of Transmission of Novel Coronavirus in. 2020. J Clin Med 9(2): 330.
- Mb WW, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus ( 2019 ‐ nCoV ) in Wuhan , China 92(4): 441-447.
- Bassetti M, Vena A, Giacobbe DR (2020) The novel Chinese coronavirus ( 2019-nCoV ) infections : Challenges for fighting the storm 50(3): e13209.
- WHO (2020) Novel Coronavirus (2019-nCoV ) Situation Report-11.
- WHO (2020) Coronavirus Disease 2019. Situation Report - 40. WHO (2020) Coronavirus disease (COVID-19) Situation Report. 71.
- WHO (2020) Coronavirus disease 2019 ( COVID-19 ) Situation Report - 101.



