Jin-Ichi Sasaki*, Chiaki Yanagihara, Hiromi Ohashi, Yasuko Kimura, C Shinohara, H Soga, Y Itoh
Dept. Bioscience and Laboratory Medicine, Hirosaki University Graduate School of Health Sciences, 66-1 Honcho, Hirosaki 036-8564, Japan
*Corresponding Author: Jin-Ichi Sasaki, Dept. Bioscience and Laboratory Medicine, Hirosaki University Graduate School of Health Sciences, 66-1 Honcho, Hirosaki 036-8564, Japan.
Abstract
Survival of enterohemorrhagic E. coli O157:H7 was studied in a water environment to know behaviours in a nutrient-poor environment. Initial cell number at 108/mL kept alive till 3 years, even though cells decreased to 104/mL, whereas control E. coli died out 2.5 years in this condition. Three years survived cells grew as colourless colonies on Rainbow Agar, but they (50 colonies) were all agglutinated with O157:H7 antiserum. Water survived cells showed elongated forms with a surface smooth in scanning electron microscopic view. Whereas control O157:H7 cells that maintained on nutrient agar produced black colonies, and cells length were 2-3μm. Survived cells were less Stx producibility. On Stx IP animal tests O157:H7 toxin bound to liver and kidney cells, along with severe haemorrhagic intestine.
Keywords: enterohaemorrhagic E. coli O157:H7, survival in water, colony appearance on Rainbow Agar, elongation, Stx producibility, Stx binding in liver and kidney cells, heat shock protein (HSP), viable but non-culturable (VBNC)
Introduction
Hemorrhage E. coil O157:H7 was first reported as a pathogen in 1982 during an outbreak investigation of hemorrhagic colitis [1]. The first unprecedented massive outbreak caused O157:H7 in Japan was occurred in May 1996 at the elementary school Okayama Prefecture south part of Japan, including 486 school students and two deaths [2]. The onset of EHEC 0157:H7 infection is bloody diarrheal stool and proceeds to the development of hemolytic uremic syndrome (HUS) caused by Stx toxin, and some of them are led to death. Thereafter EHEC 0157:H7 infection was gradually moving up to the north and ended at Hokkaido in autumn leaving big sacrifices in Japan. Health Center inspection made reports that the bean sprout salad served in school lunch was the most suspicious causative agent, but definitive evidence was not obtained [3].
EHEC 0157:H7 caused food poisoning was very rare till then in Japan. After this historical large outbreak, EHEC 0157:H7 food poisonings were successively reported each year in our country, suggesting EHEC 0157:H7 agent settled down and contaminated in nationwide area, but we lacked information of O157:H7 behaviours how they were surviving in a natural environment.
Under these situations experiments were designed to focus on EHEC 0157:H7 behaviours in water environment to gain fundamental knowledge, comparing to non-pathogenic Escherichia coli.
2. Materials and Methods
2.1 Bacteria and Survival Test in Water Environment
Enterohemorrhagic E. coli 0157:H7 and non-pathogen Escherichia coli isolated from the patient and health person were kindly provided by the Aomori Prefectural Health Research Centre in Japan. Both bacteria were overnight incubated in Nutrient broth (Oxoid, England) and each 0.1mL was suspended in 10mL distilled water and kept in the experiment room to count viable cells at the planned intervals. Grown colonies on Rainbow agar plate were tested antigenicity against anti-EHEC O157:H7 antibody (Denka, Japan) each time to see antigenic changes during in water environment.
2.2 Rainbow Agar (Biology, Inc USA)
On Rainbow, Agar O157:H7 has both selective and chromogenic properties that make it particularly useful for isolating pathogenic E. coli strains. The medium contains chromogenic substrates that are specific for two E. coli -associated enzymes: ßgalactosidase (a blue-black chromogenic substrate) and ß-glucuronidase (a red chromogenic substrate). Rainbow Agar O157:H7 is listed in FDA's Bacteriological Analytical Manual (BAM) and presents the agency's preferred laboratory procedures for microbiological analyses of foods and cosmetics
EXPECTED RESULTS; (www.biology.com/.../00A-017-rA-Rainbow-Agar-Broch...)
Organism Colony Colour
E. coli O157:H7 Black or Gray
E. coli O157:H7 (glucuronidase+) Purple blue
E. coli O26:H11 Purple magenta E. coli
O48:H21 Purple
E. coli O111:H or O111:H8 Violet or Gray
2.3 Shiga Toxin (Stx, VT) Measurement
Reverse passive latex agglutination test (RPLA test) was performed to examine Stx producibility of EHEC O157:H7 maintained 3 years in a water environment using an Stx Kit (Denka, Japan). Briefly three colonies of EHEC O157: H7 grown on Rainbow Agar were transfer into 10 ml nutrient broth for overnight cultivation, and the supernatant was prepared by 3,000rpm, 15min centrifugation for Stx toxin tests.
2.4 Preparation of Stx for Animal Toxicity Test
EHEC O157:H7 cultivated in nutrient broth (UK) overnight was treated with Polymyxin B to kill bacteria and centrifuged at 3,000 rpm 10 min followed by filtration by mill pore filter for tests (undiluted solution).
2.4 Animal Tests
Three female BALB/c mice housed in a cage were administrated 1.0 mL Stx via two ways of oral (PO) and peritoneal (IP) routes. Clinical symptoms (findings) were recorded each day.
For these animal experiments, we got approval from The Ethics Committee of Hirosaki University Animal Research Organization.
2.5 Stx Detection in Liver and Kidney in Stx IP Animal Organs
Indirect immunofluorescent tests were applied to the liver and kidney sections prepared from the Stx ip animals.
2.6 Scanning Electron Microscopic Analysis of O157:H7 Cells
EHEC O157: H7 were fixed overnight in 3.0 % glutaraldehyde, followed by ethanol dehydration, isoamyl alcohol immersion, and gold-palladium deposition for scanning electron microscopic observation (JSM 5310, Japan).
3. Results
3.1 Survival Period of EHEC O157:H7 in Water Environment
Survival terms of EHEC O157 in a water environment were summarized in Table 1. Cells kept at a high rate of survival for six months, thereafter, gradually decreasing and 3 years later it became at 1.5×104 cfu/ml. Whereas control E coli died out at 2 years in water environment condition.
Table 1: Duration of survival of EHEC 0157:H7 in water environment
|
Period (Years) |
EHECO157 cfu/ml |
E. coli cfu/ml |
|
0 |
5.7×108 |
4.0×109 |
|
0.5 |
5.0×108 |
8.0×106 |
|
1.0 |
2.0×107 |
5.0×105 |
|
1.5 |
2.5×107 |
4.0×103 |
|
2.0 |
1.4×107 |
4.0×103 |
|
2.5 |
4.0×106 |
0 |
|
3.0 |
1.5×104 |
0 |
3.2 Colony Appearance on Rainbow Agar
Three years survived EHEC 0157:H7 produced less gray (gray-less) or faint gray-colored colonies on Rainbow Agar (Photo 1 A), looked different bacteria appearance, and 50 colonies were randomly selected for agglutination tests with anti-O157:H7 antibody. As all colonies tested reacted with antibodies and were identified as EHEC O157:H7, indicating antigen stability even in less nutritious water conditions.

Photo 1. Colonies appearance on Rainbow Agar of 3 years survived EHEC O157:H7 in water environment
(A); EHEC O157:H7 colonies survived 3 years in a water environment. Colonies were mostly colourless or faint.
(B); Nutrient agar maintained EHEC O157:H7 (Control)
3.3 Stx Producibility of 3 Years Survived EHEC O157:H7 in Water Environment
Three years survived O157:H7 in water weakened in toxin producibility, and attenuation liability was prominent in Stx 2 producibility (Table 2).
Table 2: Stx producibility of 3 years survived EHEC 0157:H7
|
EHEC0157(Maintained in) |
Colony No. |
Stx1(Dilution) |
Stx2 (Dilution) |
|
Water |
1 |
8 |
32 |
|
2 |
8 |
64 |
|
|
3 |
16 |
8 |
|
|
Nutrient agar |
1 |
64 |
128< |
|
2 |
32 |
128< |
|
|
3 |
16 |
128< |
3.4 Stx Toxicity Test against Balb/c Mice by Two Different Ways IP and Oral Survival days of Stx administered mice via IP or oral
|
Stx (refer to 2.4) |
Days Survived |
|
Undiluted, IP |
1.5 |
|
100×Diluted, IP |
3.5 |
|
Undiluted, Oral |
5.0< |
|
100×Diluted, Oral |
5.0< |
|
80℃, 30min, IP |
5.0< |
IP administration demonstrated the highest lethal toxicity against mice. Whereas the oral route was less toxic to animals, and the Stx activity was destroyed by heat treatment at 80℃, 30 min.
3.5 Stx IP Mouse Peritoneal Cavity and Haemorrhagic Intestine
Clinical findings of Stx IP Balb/c mice; bloody rheum, respiration disorder, loss of appetite, lower body temperature, tetraplegia, and convulsion
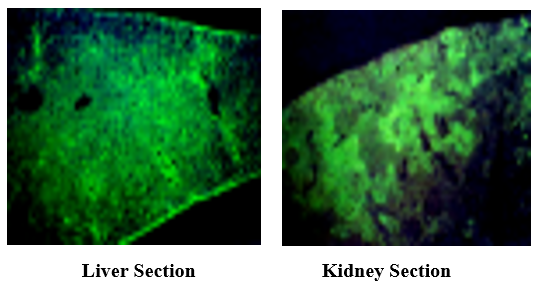
3.6 Toxin Detection in Stx IP Mice Liver and Kidney by Indirect Immuno-Fluorescence Analysis
Stx has mainly detected Glissons’ capsule and hepatocytes with intense fluorescence (left, cv: central vein), and the renal corpuscles in the kidney (arrow) produced strong fluorescence (right).
3.7 Elongated Cells of 3 Years Survived EHEC O157:H7 in Water Environment
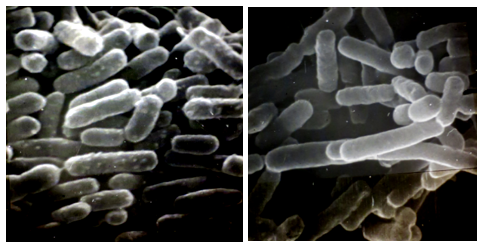
Photo 2: Scanning electron micrographs of EHEC O157:H7 survived 3 years in a water environment (right), and nutrient agar maintained EHEC O156:H7 (left).
Bar indicates 1μm.
EHEC O157:H7 in a water environment showed elongated cells with smooth surfaces as seen in the picture (right). Probably less nutrition affected the cell's division and surface structures.
Discussion
EHEC 0157: H7 was first recognized as a pathogen in 1982 during an outbreak investigation of haemorrhagic colitis EHEC O157 infection can lead to haemolytic uremic syndrome (HUS), characterized by haemolytic anaemia, thrombocytopenia, and renal injury. To understand Stx pathogen to kidney including other organs toxins were IP injected and mainly kidney and liver were pathologically inspected to know whether Stx bound to these cells to make damages.
On May 28, 1996, an outbreak of food poisoning due to EHEC O157:H7 arose in the primary school in Okayama Prefecture Japan, 468 patients and 2 died caused by HUS. EHEC O157:H7 caused food poisoning serially occurred at elementary school via school lunch and almost came to end in Autumn at Hokkaido. After this historical huge outbreak, sporadic cases were reported each year that was never experienced before, suggesting this bacterium passed over the cold winter. To fight pathogens there are two strategies, collecting enemy information and developing weapons. As a weapon, we had developed the silver-coated ceramic applied oligodynamic action of silver in water [4].
At that stage, we shared less information on what going on with EHEC O157:H7 in water, soil, vegetables, hence mimicked less nutritious water environment was prepared to trace O157:H7 behaviours. In this situation O157:H7 showed us different behaviour on Rainbow agar forming colourless colonies, lowering Stx 1 and 2 producibilities, and cell elongation with a smooth surface. Colourless these colonies reacted with EHEC 0157:H7 antibody, and antigenicity was stable in a water environment. Therefore, agglutination test was essential for identification, and Rainbow agar observation was used just as a reference. When used Rainbow agar for O157:H7 isolation from water, they reported no 0157:H7 in inspected water because of all colourless colonies, and someone misunderstood that O157:H7 entered a viable but non-culturable (VBNC) state. The further report mentioned that survival of O157:H7 in water was mainly affected by the outer membrane protein composition, but not change in lipopolysaccharide and O157:H7 is a hardy pathogen that can survive for long period in water [5]. The outer membrane on bacteria is just a protein layer to protect against toxic substances invasion and is easily deleted.
The bacterial elongation phenomenon is occasionally observed as a kind of natural mutation caused to the failure of cell division. In our past studies on toxin producibility of Clostridium botulinum, a single cell from one colony was picked up by the manipulator and cultivated to check toxin production. A wide variety of toxin producibility in single-cell isolated was observed [6]. In other words, one colony consisted of a different nature single cell. This experimental fact raises the following question, where is the border between mutation and selection.
In response to unsuitable conditions like starvation or stress bacteria change their metabolic activity and cellular components to maintain viability to extend bacterial survival, and some cases enter the non-growing stage, and the heat shock protein(s) (HSP 60 and 69 kDa) with viable but non-culturable (VBNC) appeared associated each other to bacterial survival, therefore designing thermal processes in beef gravy must be considered for microbiological safety of thermally processed foods [7].
Reference: Shape of Mycobacterium BCG HSP 65 kDa (GroEL) electron-micrographs (left and centre), and HSP model (right). HSP size 20nm.
Filled up peptides for folding in centre hole (?)
The 65 kDa HPS is considered to associate with varieties of bio-functional roles as tumour antigens, widely detected in animals and human tumour cells, leading to cancer immunotherapy studies [8], host defence against various microbial pathogens, and autoimmune inflammatory disorders [9]. HSP probably has a cross-talking ability with stress burden and VBNC in the microbiology world in nature and providing fantastic research subjects in the future.
Conclusion
Finding results obtained in this experiment were.
· EHEC O157:H7 survived over 3 years in a less nutritional water environment.
· Survived bacteria in water produced colourless or faint-coloured colonies on Rainbow
Agar, in which standard O157:H7 bacteria formed black or gray colonies. Fifty colonies positively reacted with anti-EHEC O157:H7 monoclonal antibody.
· Stx producibility also weakens, and toxin amount produced lowed.
· In Stx IP injected mouse toxin was detected in liver and kidney by indirect immunofluorescence.
· EHEC O157:H7 in water elongated with a smooth surface.
Competing Interests: Authors have declared no competing interests exist.
Reference
- JM Rangel, PH Sparling, C Crowe, PM Griffin, DL Swerdlow (2005) Epidemiology of Escherichia coli O157:H7 Outbreaks, United States 1982-2002. Emerging Infectious Diseases. 11(4): 603–609.
- IASR: The Topic of This Month Vol. 17 No. 8 (No. 198), Outbreaks of hemorrhagic Escherichia O157:H7 infection, 1996, Japan
- Fukushima H, Hashizume T, Morita Y, Tanaka J, Azuma K, et al. (1999) Clinical experiences in Sakai City Hospital during the massive outbreak of enterohemorrhagic Escherichia coli O157 infections in Sakai City, 1996. Pediatrics International. 41(2): 213-217
- J Sasaki, T Kita, N Kurota, T Yoshisawa: Development of anti-enterohemorrhagic E. coli O157 ceramic. Clinical and Experimental Medicine (in Japanese), Vol 187(2): 157-158, 1998
- G wang, MP Doyle (1998) Survival of enterohemorrhagic Escherichia coli O157:H7 in Water. J Food Prot. 61(6) :662-7.
- J Sasaki: Studies on the toxigenic variation of Clostridium botulinum type E using micro-manipulator. Hirosaki Medical J. 27;431-440, 1975
- Juneja VK, Klein PG, Marmer BS (1998) Heat shock and thermotolerance of E coli O157:H7 in a model beef gravy system and ground beef. J Applied Microbiology. 84(4): 677-684.
- Sasaki J, Dejehansart M, De Bruyn J (1994) The expression of mycobacterial heat shock protein (HSP64) on Meth A tumor cells. Immunol Cell Biol. 72(5): 415-418.
- Sakai T, Hisaeda H, Ishikawa H, Maekawa Y, Zhang M, et al. (1999) Expression and role of heat-shock protein 65 (HSP 65) in macrophages during Trypanosoma cruzi infection: involvement of HSP 65 in the prevention of apoptosis of macrophages. Microbes Infect. 1(6): 419-27.