Khatija1, Muhammad Aetesam Nasir2, Falak Sher2, Ifrah Saroosh2, Aisha Shakir2, Muhammad Abdullah2, Gull Zaman3, Hira Ashiq3, Muhammad Waqar Mazhar4*
1Department of Microbiology, Government College University, 38000 Faisalabad, Pakistan.
2Hitec Institute of Medical Sciences Taxila Cantt Rawalpindi, Pakistan.
3University of Health Sciences Lahore, Medicine Lahore, Punjab, Pakistan.
4Department of Bioinformatics and Biotechnology, Government College University, 38000 Faisalabad, Pakistan.
*Corresponding Author: Muhammad Waqar Mazhar, Department of Bioinformatics and Biotechnology, Government College University, 38000 Faisalabad, Pakistan.
Abstract
Background: Antimicrobial-resistant (AMR) pathogens causing Urinary Tract infection is a severe public health concern in our clinical setting.
Methodology: Therefore, the current study was designed to investigate AMR profiles and the prevalence of bacterial pathogens in catheterized pediatric patients. 200 catheter tips were collected from the different wards (medical, surgical, urology) at the Children's Hospital Faisalabad. Samples were streaked on nutrient agar plates, and the positivity of the samples was noted after 24 hours. Positive samples were processed further to identify K. pneumonia, P. aeruginosa, S. aureus, and E. coli using culture identification, microscopy, and biochemical profiling based on culture characterization, biochemical profiling, and antibiotic susceptibility testing.
Results: 76 (38%) samples showed growth on nutrient agar. In processed samples, the high prevalence was marked for P. aeruginosa (24/200; 12%) followed by E. coli (22/200; 11%) and S. aureus (19/200; 9.5%), while 11 K. pneumoniae isolates (5.5%) were identified in this study. In antibiotic susceptibility profiling of P. aeruginosa, the highest susceptibility was found for colistin (100%) and imipenem (70.83%), followed by gentamicin (54.17%), while the highest resistance was found for tobramycin (54.17%) followed by meropenem, ceftazidime, and cefotaxime (50%). In antibiotic susceptibility profiling of K. pneumonia, the highest susceptibility was found for colistin (100%) and imipenem (72.73%), followed by gentamicin and ciprofloxacin (45.45%), while the highest resistance was found for cefotaxime (63.63%) followed by meropenem, tobramycin, and amikacin (54.54%). In antibiotic susceptibility profiling of E. coli, the highest susceptibility was found for colistin (100%) and imipenem (63.64%), followed by ciprofloxacin (54.55%) while the highest resistance was found for gentamicin (54.55%) followed by tobramycin, meropenem, ceftazidime, and amikacin (50%).
Conclusion: In antibiotic susceptibility profiling of S. aureus, the highest susceptibility was found for vancomycin (100%), clindamycin, cefoxitin, and trimethoprim-sulfamethoxazole (57.89%), while the highest resistance was found for erythromycin and ampicillin (47.37%).
Conclusion: Advance studies are needed to investigate the actual investigations of bacterial contamination; resistance to treatment options and antibiotics are required.
Keywords: Antibiogram; Cather-associated bacteria; urinary tract infection; Antibiotic Resistance
Introduction
Background
In children, urinary tract infection (UTI) is the most prevalent bacterial infection within the first seven years of life, affecting 8% and 2% of girls and boys, respectively [8]. Abnormalities of urinary tract abnormalities, like congenital, can cause a high risk of UTI in some children [6]. In 30% of children with CAKUT (congenital anomalies of kidney and urinary tract) are at danger for the development of UTI in children. Unidirectional flow of urine changes due to vesico-ureteral reflux (VUR) [11], while pyelo-ureteral junction obstruction (PUJO) leads to stasis, in which both increase the risk of multiplying pathogenic microorganisms [4]. At the age of 1 month and 11 years, more than 8% of children will experience at least one UTI, and during the first six to 12 months after an initial UTI, more than 30% of kids and newborns experience repetitive infections [3]. The most common etiology of UTIs is due to more than 95% of bacteria. Escherichia coli (E. coli) is the most frequent causative organism of UTIs and is responsible for more than 80% [14]. In males, Proteus mirabilis is more frequent than in females, while in newborn infants, Streptococcus agalactiae is more common, Streptococcus viridians, Haemophilus influenza, Streptococcus pneumonia, Staphylococcus epidermidis, Staphylococcus aureus, and Streptococcus agalactia may be responsible in children with anomalies of the urinary tract (anatomic, neurologic, or functional) or compromised immune system [13]. Only a proper identification of the local pathogen and information on the susceptibility patterns and any related risk factors can provide appropriate treatment for UTIs [14]. Because of incorrect antibiotic use, the bacterial sensitivity pattern of common pathogens is gradually changing in all countries [15]. To decrease the morbidity rate of UTIs, proper treatment is required. The non-specific signs and symptoms of UTIs in children under the age of two years can make it challenging to diagnose UTIs [2]. Children with simple UTIs may respond to sulphonamides, amoxicillin, trimethoprim-sulfamethoxazole, or cephalosporins, with amoxicillin, sulphonamides, trimethoprim-sulfamethoxazole, or cephalosporins concentrating in the lower urinary tract [19]. In high-income countries suggest that bacteria that cause UTIs are more likely to form resistance to conventional antibiotics such as trimethoprim-sulfamethoxazole [16]. The fatality rate of S. aureus has been minimized with the help of antibiotics, but S. aureus quickly develops resistance to antibiotics. Factors like toxins, adhering proteins, enzymes, antimicrobial peptides, and super-antigen make it a significant pathogen for humans and animals [18]. Multidrug-resistant Escherichia coli has been a topic of concern in the current era because of its wide host range, elevation in its pathogenicity level, competency in survival, and many reported pandemics [5]. Multidrug resistance (MDR) in E. coli is a serious issue that poses a risk to human and animal health [1].
This study aims to collect and identify the isolates recovered from the clinical specimens from pediatric patients and the antimicrobial resistance of bacterial isolates as per CLIC guideline 2020.
Material and Methodology
Ethical Consideration
Before starting the study, ethical permission was obtained from the Ethical Review Committee, Government College University Faisalabad.
Consent Forms
A consent form was designed that included name, gender, date and time of sampling, and permission from the patients/guardians to use their samples for research purposes. Consent forms were filled out by the patients/guardians at the time of the sampling. The data of the patients were kept secret and not shared with anyone.
Sample Collection
200 catheter tips were collected from the pediatric patients of different wards (urology, surgery, medicine) at the Children's Hospital Faisalabad. The clinical samples of catheter tips were collected using sterile scissors and cutting catheter tips from the balloon side by 2cm and transferred into a clean container.
Sample Processing and Staining
Samples were first kept in pre-prepared nutrient broth for 24 hours. The Broth was subcultured on Blood, nutrient, and MacConkey agar plates and incubation were done at 37 ºC overnight. Bacterial isolate colonies were preliminarily identified based on colony morphology, the isolates' color pigment, and the colonies' size and shape.
Gram Staining
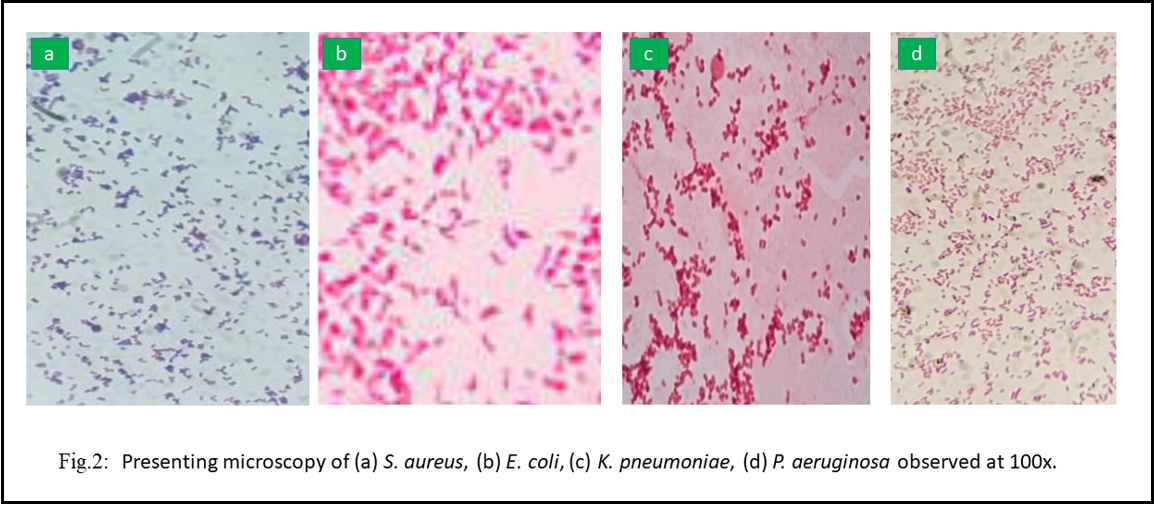
The basic principle of gram staining is distinguishing between gram-positive and gram-negative bacteria based on a cell wall. Gram staining of the isolates included smear preparation, Gram staining, and microscopy of the colonies. The gram staining was observed at 100x under the microscope; Gram-positive isolates appear purple-blue, while Gram-negative isolates appear pink.
Biochemical Profiling Of Isolates
Isolates were processed further for biochemical Profiling for confirmation of biochemical characteristics. Oxidase, triple sugar iron, citrate, urease, indole, methyl red, and Voges Proskauer tests were conducted, and results were noted for each of the processed isolates.
Antibiotic Susceptibility Testing
Hudzicki & Kirby-Bauer,2016 method measured the sensitivity of bacteria. Results were recorded while different zones appeared on antibiotic agar plates.
Statistical Analysis
Data were analyzed by SPSS software; sheets were prepared for each tested sample. Statistical interpretations were performed for analysis of the results.
Results
This study processed two hundred samples, and 76 (38%) showed growth on nutrient agar. Sample positivity has been presented in (Table 1. Table 2) Sample positivity for the tested samples.
Table 1: Demographic distribution of total patients
|
Demographic characteristic |
Category |
Number (n) |
Percentage (%) |
|
Gender |
Male |
120 |
60% |
|
Female |
80 |
40% |
|
|
Male to female ratio |
3:2 |
||
|
Sample distribution |
Medical ward |
68 |
34% |
|
Surgical ward |
66 |
33% |
|
|
Urology ward |
66 |
33% |
|
Table 2: Positive and negative samples distribution
|
|
Frequency (n) |
Percentage % |
|
Total samples |
200 |
- |
|
Positive samples |
76 |
38% |
|
Negative samples |
124 |
62% |
Prevalence of bacteria in samples76 samples marked positive were processed further to estimate the prevalence of bacteria. In processed samples, high prevalence was observed for P. aeruginosa (24/200; 12%) followed by E. coli (22/200; 11%) and S. aureus (19/200; 9.5%), while 11 K. pneumoniae isolates (5.5%) were identified in this study. The results for the prevalence of bacteria have been presented in (Table 3).
Table 3: Prevalence of bacteria in samples
|
Bacterial specie |
Frequency (n) |
Prevalence |
|
P. aeruginosa |
24 |
12% |
|
E. coli |
22 |
11% |
|
S. aureus |
19 |
9.5% |
|
K. pneumonia |
11 |
5.5% |
Patients’ clinical demographic distribution for P. aeruginosa
In a study of demographic factors for P. aeruginosa, in overall sample distribution for investigation of gender, the high prevalence was found for males (15%), of inquiry of sample location, the high majority was found for surgical wards and urological wards (12.12%), and for analysis of age group, the high majority was found for age group 1-4 (13.54%). The results for the patient's clinical demographic distribution for P. aeruginosa have been presented in (Table 4).
Table 4: Patients clinical demographic distribution for P. aeruginosa
|
Demographic factor
|
Category |
No. of samples
|
Frequency of P. aeruginosa |
Prevalence |
|
Gender
|
Male |
120 |
18 |
15% |
|
Female |
80 |
06 |
7.5% |
|
|
Sample location |
Medical Ward |
68 |
08 |
11.76% |
|
Surgical Ward |
66 |
08 |
12.12% |
|
|
Urology Ward |
66 |
08 |
12.12% |
|
|
Age group (years) |
01-04 |
96 |
13 |
13.54% |
|
05-08 |
64 |
06 |
9.38% |
|
|
09-12 |
40 |
05 |
12.5% |
Patient's clinical demographic distribution for E. coli
In the study of demographic factors for E. coli, in overall sample distribution for investigation of gender, a high prevalence was found for males (13.33%). For analysis of sample location, a high majority was found for urological wards (18.18%), and for investigation of age group, a high prevalence was found for age group 5-9 (10.94%). The results for patients’ clinical demographic distribution for E. coli have been presented in (Table 5).
Table 5: Patients clinical demographic distribution for E. coli
|
Demographic factor |
Category |
No. of samples |
Frequency of E. coli |
Prevalence |
|
Gender |
Male |
120 |
16 |
13.33% |
|
Female |
80 |
06 |
7.50% |
|
|
Sample location |
Medical ward |
68 |
03 |
4.41% |
|
Surgical ward |
66 |
07 |
10.29% |
|
|
Urology ward |
66 |
12 |
18.81% |
|
|
Age group (years) |
01-04 |
96 |
11 |
11.46% |
|
05-08 |
64 |
07 |
10.94% |
|
|
09-12 |
40 |
04 |
10% |
Patient's clinical demographic distribution for K. pneumoniae
In the study of demographic factors for K. pneumoniae, in the overall sample distribution for investigation of gender, a high prevalence was found for males (7.5%). For analysis of sample location, a high majority was found for surgical wards (6.06%), and for investigation of age group, high prevalence was found for age group 1-4 (7.30%). The results for the patient's clinical demographic distribution for K. pneumoniae have been presented in (Table 6).
Table 6: Patients clinical demographic distribution for K. pneumoniae
|
Demographic factor |
Category |
No. of samples |
Frequency of K. pneumonia |
Prevalence |
|
Gender |
Male |
120 |
09 |
7.50% |
|
Female |
80 |
02 |
2.50% |
|
|
Sample location |
Medical ward |
68 |
04 |
5.88% |
|
Surgical ward |
66 |
04 |
6.06% |
|
|
Urology ward |
66 |
03 |
4.55% |
|
|
Age group (years) |
01-04 |
96 |
07 |
7.30% |
|
05-08 |
64 |
02 |
3.13% |
|
|
09-12 |
40 |
02 |
5% |
Patients’ clinical demographic distribution for S. aureus
In the study of demographic factors for S. aureus, in the overall sample distribution for investigation of gender, a high prevalence was found for males (11.67%). For analysis of sample location, a high majority was found for surgical wards (10.61%), and for analysis of age group, a high majority was found for age group 1-4 (14.58%). The results for the patient's clinical demographic distribution for S. aureus have been presented in Table 7.
Table 7: Patients clinical demographic distribution for S. aureus
|
Demographic factor |
Category |
No. of samples |
Frequency of S. aureus |
Prevalence |
|
Gender |
Male |
120 |
14 |
11.67% |
|
Female |
80 |
05 |
6.25% |
|
|
Sample location |
Medical ward |
68 |
06 |
8.82% |
|
Surgical ward |
66 |
07 |
10.61% |
|
|
Urology ward |
66 |
06 |
9.09% |
|
|
Age group (years) |
01-04 |
96 |
14 |
14.58% |
|
05-08 |
64 |
03 |
4.69% |
|
|
09-12 |
40 |
02 |
5% |
Confirmation of the Isolates
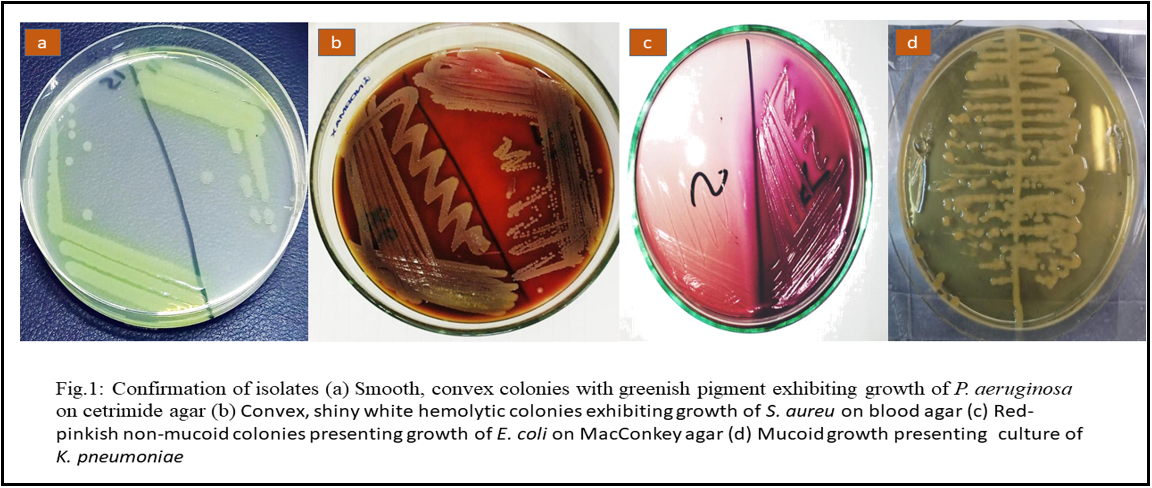
For confirmation of the isolates, identification of P. aeruginosa was carried out on cetrimide agar, and smooth, convex colonies with greenish pigment and grape-like odor are the characteristic features of the P. aeruginosa isolates. Identifying E. coli was carried out on MacConkey agar to confirm that the isolates and red-pinkish non-mucoid colonies are distinctive features for E. coli separates. For the confirmation of the isolates, identification of S. aureus was carried on blood agar and convex, shiny white hemolytic colonies are characteristic features for S. aureus isolates. Identifying K. pneumoniae was carried out on EMB agar to confirm the isolates and mucoid pinkish growth is a distinctive feature of K. pneumoniae isolates. Development exhibiting the culture characteristics of P. aeruginosa, E. coli, K. pneumonia, and S. aureus has been presented in Figure 1. The isolates were observed under microscope 100 X, shown in Figure 2.
Biochemical profiling of isolates
Biochemical profiling of the isolates was carried out for the confirmation of the biochemical characteristics of the isolates. The results of the biochemical profiling of the isolates have been presented in (Tables 8 and 9).
Table 8: Biochemical profiling for Gram negative isolates
|
Bacteria |
Oxidase |
TSI |
Indole |
Citrate |
Urease |
Methyl red |
Voges Proskaue r |
|
E. coli |
Negative |
Positive |
Positive |
Negative |
Negative |
Positive |
Negative |
|
P. aeruginosa |
Positive |
Negative |
Negative |
Positive |
Negative |
Negative |
Negative |
|
K. pneumonia |
Negative |
Positive |
Negative |
Positive |
Positive |
Negative |
Positive |
Table 9: Biochemical profiling for S. aureus isolates
|
Catalase |
Coagulase |
|
Positive |
Positive |
Antibiotic susceptibility testing
Antibiotic susceptibility testing was conducted against the enlisted antibiotics, and results were formulated according to the CLSI 2021 guidelines. The results of antibiotic susceptibility profiling of the isolates have been presented in (Table 10-13).
Table 10: Presenting antibiotic susceptibility profiling of P. aeruginosa isolates.
|
Antibiotic |
Susceptible |
Intermediate |
Resistant |
|
Gentamicin |
13 (54.17%) |
02 (8.33%) |
09 (37.50%) |
|
Ciprofloxacin |
12 (50%) |
01 (4.17%) |
11 (45.83%) |
|
Meropenem |
09 (37.50%) |
03 (12.50%) |
12 (50%) |
|
Imipenem |
17 (70.83%) |
02(8.33%) |
05 (20.83%) |
|
Tobramycin |
10 (41.67%) |
01(4.17%) |
13 (54.17%) |
|
Ceftazidime |
11 (45.83%) |
01(4.17%) |
12 (50%) |
|
Cefotaxime |
10 (41.67%) |
02(8.33%) |
12 (50%) |
|
Amikacin |
12 (50%) |
01(4.17%) |
11 (45.83%) |
|
Colistin |
24 (100%) |
0 |
0 |
|
Ampicillin |
0 |
0 |
100 |
Table 11: Presenting antibiotic susceptibility profiling of K. pneumoniae isolates.
|
Antibiotic |
Susceptible |
Intermediate |
Resistant |
|
Gentamicin |
05 (45.45%) |
01 (9.09%) |
05 (45.45%) |
|
Ciprofloxacin |
05 (45.45%) |
02 (18.18%) |
04 (36.36%) |
|
Meropenem |
04 (36.36%) |
01 (9.09%) |
06 (54.54%) |
|
Imipenem |
08 (72.73%) |
0 |
03 (27.27%) |
|
Tobramycin |
04 (36.36%) |
01 (9.09%) |
06 (54.54%) |
|
Ceftazidime |
04 (36.36%) |
01 (9.09%) |
06 (54.54%) |
|
Cefotaxime |
03 (27.27%) |
01 (9.09%) |
07 (63.63%) |
|
Amikacin |
04 (36.36%) |
01 (9.09%) |
06 (54.54%) |
|
Colistin |
11 (100%) |
0 |
0 |
|
Ampicillin |
0 |
0 |
100 |
Table 12: Presenting antibiotic susceptibility profiling of E. coli isolates.
|
Antibiotic |
Susceptible |
Intermediate |
Resistant |
|
Gentamicin |
09 (40.91%) |
01 (4.55%) |
12 (54.55%) |
|
Ciprofloxacin |
12 (54.55%) |
02 (9.10%) |
08 (36.36%) |
|
Meropenem |
09 (40.91%) |
02 (9.10%) |
11 (50%) |
|
Imipenem |
14 (63.64%) |
01 (4.55%) |
07 (31.82%) |
|
Tobramycin |
09 (40.91%) |
02 (9.10%) |
11 (50%) |
|
Ceftazidime |
08 (36.36%) |
03 (13.64%) |
11 (50%) |
|
Cefotaxime |
10 (45.45%) |
02 (9.10%) |
10 (45.45%) |
|
Amikacin |
10 (45.45%) |
01 (4.55%) |
11 (50%) |
|
Colistin |
22 (100%) |
0 |
0 |
|
Ampicillin |
0 |
0 |
100 |
Table. 13: Presenting antibiotic susceptibility profiling of S. aureus isolates
|
Antibiotic |
Susceptible |
Intermediate |
Resistant |
|
Penicillin |
08 (42.11%) |
03 (15.79%) |
08 (42.11%) |
|
Cefoxitin |
11 (57.89%) |
01 (5.26%) |
07 (36.84%) |
|
Erythromycin |
08 (42.11%) |
02 (10.53%) |
09 (47.37%) |
|
Ampicillin |
09 (47.37%) |
01 (5.26%) |
09 (47.37%) |
|
Trimethoprim-sulfamethoxazole |
11 (57.89%) |
02 (10.53%) |
06 (31.58%) |
|
Tetracycline |
09 (47.37%) |
02 (10.53%) |
08 (42.11%) |
|
Azithromycin |
08 (42.11%) |
03 (15.79%) |
08 (42.11%) |
|
Clindamycin |
11 (57.89%) |
01 (5.26%) |
07 (36.84%) |
|
Ciprofloxacin |
10 (52.63%) |
03 (15.79%) |
06 (31.58%) |
|
Vancomycin |
19 (100%) |
0 |
0 |
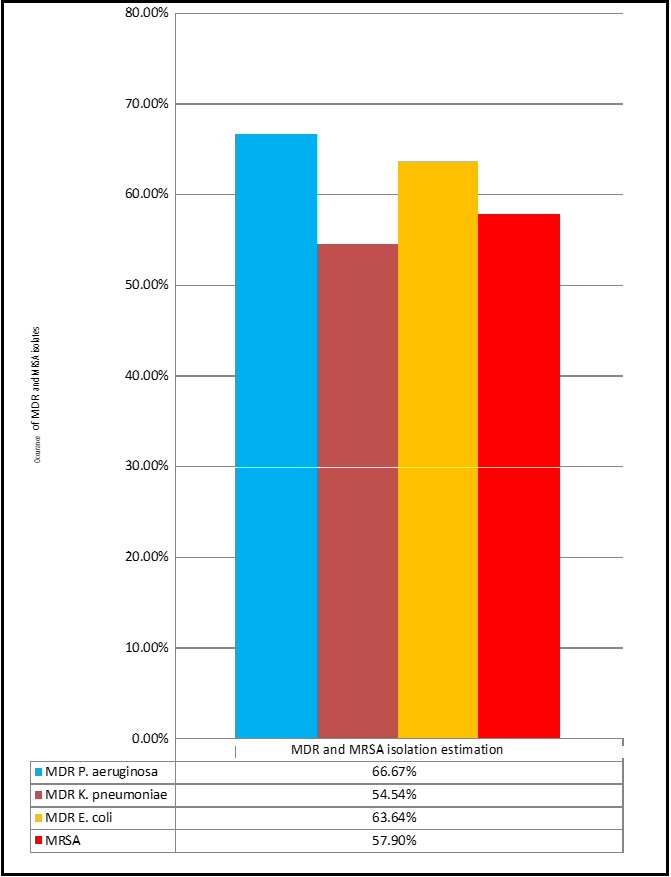
MDR and MRSA isolate estimation
For Gram-negative bacteria, the occurrence of MDR isolates was formulated based on resistance in studied isolates, while phenotypic detection of MRSA isolates was estimated by cefoxitin disk analysis.
Table 14: MDR isolates detection for studied bacteria.
|
Bacteria |
No. of isolates |
Frequency of MDR isolates |
Percentage of MDR isolates |
|
P. aeruginosa |
24 |
16 |
66.67% |
|
K. pneumoniae |
11 |
06 |
54.54% |
|
E. coli |
2 |
14 |
63.64% |
|
S. aureus |
|||
|
MRSA |
No. of isolates |
Frequency of MRSA |
Percentage of MDR isolates |
|
19 |
11 |
57.90% |
|
Figure 3: Graph presenting MDR isolates detection for studied bacteria.
Discussion
The most frequent bacterial infection in children is urinary tract infection (UTI), which affects 8% of girls and 2% of boys under the age of 7.30% of people have a chance of developing a second UTI who have already developed UTI in childhood [12]. Some diseases, such as congenital anomalies of the urinary tract, put some children at a high risk of having UTIs [7]. The upper urinary tract (pyelonephritis or kidney infection) or the lower urinary tract (cystitis or bladder infection) may be affected by UTI, and it is complicated to differentiate cystitis-based clinical symptoms and indications of pyelonephritis, particularly in children and infants [10]. Proteus mirabilis is more frequent in males than in girls while in newborn infants Streptococcus agalactiae is more common than Haemophilus influenza, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus viridians, Streptococcus pneumoniae, and Streptococcus agalactiae may be responsible in children with anomalies of the urinary tract (anatomic, neurologic, or functional) or compromised immune system [10].
Only a proper identification of the local pathogen and information on the susceptibility pattern and any related risk factors can provide appropriate treatment for UTIs. Because of incorrect antibiotic use, the bacterial sensitivity pattern of common pathogens is gradually changing in all countries [9]. Proper treatment is required to decrease the morbidity rate of UTIs [17]. Keeping in view the above facts and figures and the importance of UTIs in pediatrics, the current study was designed with the objectives to isolate and identify catheter-associated bacterial pathogens in UTIs among pediatric patients and to estimate the prevalence and antibiotic susceptibility profiling of catheter-associated bacterial pathogens in UTIs among pediatric patients.
200 catheter tips were collected from the patients of different wards (surgery, urology, medicine) at the Children's Hospital Faisalabad. Samples were first kept in pre-prepared nutrient broth for 24 hours and then streaked on nutrient agar plates, and the positivity of the samples was noted after 24 hours. Positive samples were processed further to identify E. coli, K. pneumoniae, S. aureus, and. aeruginosa using culture identification, microscopy, and biochemical profiling based on culture characterization, microscopy, and biochemical profiling. Cultures were processed on selective agar, set for incubation at 37 ºC for 24 hours, and processed further for Gram-staining, microscopy, and biochemical profiling using oxidase, catalase, triple sugar iron, urease, indole, methyl red, and Voges Proskauer test. Antibiotic susceptibility testing was performed to determine the antibiotic resistance profile of each isolate by disc diffusion method. Antibiotics were selected based on clinical relevance, which belongs to different antimicrobial groups. The zone of inhibition was interpreted according to Clinical and Laboratory Standards Institute guidelines (CLSI) 2021, and isolates were determined as resistant, intermediate, and susceptible according to CLSI guideline 2021.
200 samples were processed in this study, and 76 (38%) showed growth on nutrient agar. In processed samples, the high prevalence was marked for P. aeruginosa (24/200; 12%) followed by E. coli (22/200; 11%) and S. aureus (19/200; 9.5%), while 11 K. pneumoniae isolates (5.5%) were identified in this study. This study showed relevance with the results presented by Mishra & Wadhai (2016) in research designed on P. aeruginosa in OT samples, Mohammad et al., 2017 in research designed on K. pneumoniae in OT samples, Dhom et al. (2017) in the method of analysis on E. coli in surgical sites. These results were also supported by the results presented by Sapkota et al., (2016) in the form of research on P. aeruginosa in ward samples, Baban et al. (2019) in research designed on E. coli on surgical ward samples, and Yusuf et al., (2017) in a study intended on OT samples. In a comparative study designed on clinical isolates, Habyarimana et al. (2020) reported the prevalence of P. aeruginosa at 22.50%, E. coli at 7.5%, and K. pneumoniae isolates at 15%.
In antibiotic susceptibility profiling of P. aeruginosa, the highest susceptibility was found for colistin (100%) and imipenem (70.83%), followed by gentamicin (54.17%), while the highest resistance was found for tobramycin (54.17%) followed by meropenem, ceftazidime, and cefotaxime (50%). In a comparative study designed on catheter samples in the Czech Republic, Olejnickova et al. (2014) also reported more than 90% susceptibility to colistin; however, resistance to ciprofloxacin (56.6%) and gentamicin (42.9%) and a little susceptibility to amikacin (lesser than 10%) was reported in P. aeruginosa isolates. Bizuayehu et al. (2022) in Ethiopia also designed a comparative study on catheter samples and also wrote that imipenem, as a susceptible antibiotic (85.3%), reported high resistance to ceftazidime (83.3%) and resistance to gentamicin (41.7%) and tobramycin (41.7%) were also reported in P. aeruginosa isolates. The minor difference in results might be due to the difference in the demographic location of the study.
In antibiotic susceptibility profiling of K. pneumoniae, the highest susceptibility was found for colistin (100%) and imipenem (72.73%), followed by gentamicin and ciprofloxacin (45.45%), while the highest resistance was found for cefotaxime (63.63%) followed by meropenem, tobramycin, and amikacin (54.54%). Hyun et al. (2019) designed a study on clinical samples in Korea and reported high susceptibility to amikacin (94.4%), gentamicin (80.3%), ciprofloxacin (70.4%), and cefotaxime (53.5%) were reported. The difference in results might be due to differences in sample type and location of the sampling.
In antibiotic susceptibility profiling of E. coli, the highest susceptibility was found for colistin (100%) and imipenem (63.64%), followed by ciprofloxacin (54.55%) while the highest resistance was found for gentamicin (54.55%) followed by tobramycin, meropenem, ceftazidime, and amikacin (50%). In a comparative study designed on clinical samples in Korea, Hyun et al. (2019) reported 99.2% susceptibility to amikacin, 56% to ciprofloxacin, and 66.1% to gentamicin. These results were also supported by El-Mahdy et al. (2021) in a study designed on catheter samples in Ethiopia in which 55.6% resistance to ceftazidime was reported. Almost similar results were also reported by Vidyasagar and Nagarathnamma (2018) in a study designed on E. coli isolates from catheter samples. They also said high susceptibility to imipenem (95.7%), amikacin (58.7%), and tobramycin (58.7%). Bizuayehu et al. (2022), in a study designed on catheter samples in Nepal in which 100% susceptibility to imipenem and 37.5% resistance to ceftazidime was reported; however, 100% susceptibility to meropenem and amikacin was also reported in E. coli isolates. Ndomba et al. (2022), in a study designed on catheter samples in Tanzania, also said 50.7% resistance to ceftazidime in E. coli isolates; however, resistance to gentamicin (43%) was also reported.
In antibiotic susceptibility profiling of S. aureus, the highest susceptibility was found for vancomycin (100%), clindamycin, cefoxitin, and trimethoprim-sulfamethoxazole (57.89%), while the highest resistance was found for erythromycin and ampicillin (47.37%). Vidyasagar & Nagarathnamma (2018), in a study designed on S. aureus in catheter samples, also reported 100% resistance to vancomycin; however, a little susceptibility to erythromycin (20%) and clindamycin (20%) was found in these isolates.
A high prevalence of pathogens in catheter samples has been alarming and worsened with resistant isolates that have not only been found resistant to antibiotics studied. Advanced studies are needed to investigate the actual investigations of bacterial contamination; resistance to treatment options and antibiotics are required.
Conclusion
This study concluded that the high prevalence was determined for P. aeruginosa (24/200; 12%) and E. coli (22/200; 11%). In this study, the male patients were mainly infected compared to females (3:2). The antimicrobial profile suggested that 54.17 % of P. aeruginosa were resistant to tobramycin, and the susceptible drug was colistin (100%). In antibiotic susceptibility profiling of K. pneumoniae, the highest susceptibility was found for colistin (100%), and the highest resistance was found for cefotaxime (63.63%).
In antibiotic susceptibility profiling of E. coli, the highest susceptibility was found for colistin (100%), while the highest resistance was found for gentamicin (54.55%). In antibiotic susceptibility profiling of S. aureus, the most heightened susceptibility was found for vancomycin (100%), while the highest resistance was found for erythromycin and ampicillin (47.37%). There should be public awareness of the use of antibiotics and a stoppage of irrational use of antibiotics. People should not take self-antibiotics, over-the-counter antibiotics should be banned, and continuous education on health care.
Advanced studies are needed to investigate the actual investigations of bacterial contamination, resistance to treatment options, and resistance to antibiotics.
List of abbreviations: E. coli (Escherichia coli).
Declaration: Consent for publication
Not applicable
Availability of data and materials
Availability of data and materials on request by the corresponding author
Competing Interests: The authors have no competing interest.
Funding: No funding for this research
Authors' Contributions: All authors contribute equally.
Acknowledgments: I am grateful to all those with whom I have had the pleasure to work during this and other related projects.
References
- Arbab S, Ullah H, Wang W, Zhang J (2022) Antimicrobial drug resistance against Escherichia coli and its harmful effect on animal health. Veterinary Medicine and Science. 8(4): 1780-1786.
- Aslam O, Aslam I (2022) Urinary tract infection in childhood. InnovAiT. 15(5): 280-285.
- Buonsenso D, Sodero G, Mariani F, Lazzareschi I, Proli F, et al. (2022) Comparison between Short Therapy and Standard Therapy in Pediatric Patients Hospitalized with Urinary Tract Infection: A Single Center Retrospective Analysis. Children. 9(11): 1647.
- Harper L, Blanc T, Peycelon M, Michel JL, Leclair MD, et al. (2022) Circumcision and Risk of Febrile Urinary Tract Infection in Boys with Posterior Urethral Valves: Result of the CIRCUP Randomized Trial. Eur Urol. 81(1): 64-72.
- Davies R, Wales A (2019) Antimicrobial resistance on farms: a review including biosecurity and the potential role of disinfectants in resistance selection. Comprehensive reviews in food science and food safety. 18(3): 753-774.
- Isac R, Basaca DG, Olariu IC, Stroescu RF, Ardelean AM, et al. (2021) Antibiotic resistance patterns of uropathogens causing urinary tract infections in children with congenital anomalies of kidney and urinary tract. Children. 8(7): 585.
- Isac R, Stroescu R, Olariu C, Pop M, Farkas F, et al. (2019) Diagnosis and clinical consequences of urinary tract malformation in children. Revista Societăţii Române de Chirurgie Pediatr. 22(1): 27-30.
- Kanellopoulos TA, Salakos C, Spiliopoulou I, Ellina A, Nikolakopoulou NM, et al. (2006) First urinary tract infection in neonates, infants and young children: a comparative study. Pediatric Nephrology. 21(8): 1131-7.
- Kariyappa P, Reddy SC, Mavinahalli AS, Rao US (2020) Urinary tract abnormalities in children with first urinary tract infection. Indian Journal of Child Health.7(8): 337-339.
- Leung AKC, Wong AHC, Leung AAM, Hon KL (2019) Urinary tract infection in children. Recent patents on inflammation & allergy drug discovery. 13(1): 2-18.
- Ludwikowski BM, González R (2019) Progress in Pediatric Urology in the Early 21st Century. Front. Pediatr. 7: 349.
- Millner RO, Preece J, Salvator A, McLeod DJ, Ching CB, et al. (2019) Albuminuria in pediatric neurogenic bladder: identifying an earlier marker of renal disease. Urology. 133: 199-203.
- Mirzaei R, Mohammadzadeh R, Alikhani MY, Shokri Moghadam M, Karampoor S, et al. (2020) The biofilm‐associated bacterial infections unrelated to indwelling devices. IUBMB life. 72(7): 1271-1285.
- Oumer Y, Regasa Dadi B, Seid M, Biresaw G, Manilal A (2021) Catheter-associated urinary tract infection: Incidence, associated factors and drug resistance patterns of bacterial isolates in southern ethiopia. Infection and drug resistance. 14: 2883-2894.
- Ponvelil JJ, Gowda HN, Raj SMR (2020) Prevalence of urinary tract infection and sensitivity pattern amongst children less than 3 years of age with fever in a tertiary care hospital in South Karnataka. Int J Basic Clin Pharmacol. 9(5): 736-742.
- Sangeda RZ, Paul F, Mtweve DM (2021a) Prevalence of urinary tract infections and antibiogram of uropathogens isolated from children under five attending Bagamoyo District Hospital in Tanzania: A cross-sectiona l study [version 1; peer review: awaiting peer review]. F1000. 10: 449.
- Sangeda RZ, Paul F, Mtweve DM (2021b) Prevalence of urinary tract infections and antibiogram of uropathogens isolated from children under five attending Bagamoyo District Hospital in Tanzania: A cross-sectional study. F1000Research. 10: 449.
- UMOH HP (2022) CHARACTERIZATION OF staphylococcus aureus ISOLATED FROM DOOR HANDLES IN THE COLLEGE OF HUMANITIES, MANAGEMENT AND SOCIAL SCIENCES, MOUNTAIN TOP UNIVERSITY.
- Wang ME, Greenhow TL, Lee V, Beck J, Bendel-Stenzel M, et al. (2021) Management and outcomes in children with third-generation cephalosporin-resistant urinary tract infections. Journal of the Pediatric Infectious Diseases Society: 10(5): 650- 658.